Concluding Remarks 🎉
1. What We Learned Today
We have covered the foundational pillars of R programming for bioinformatics:
The Ecosystem: Understanding the difference between Base R, RStudio, CRAN, BioConductor, and GitHub.
The Basics: Mastering data types (vectors, factors, lists) and the “home base” of the Working Directory.
Data Wrangling: Using
dplyrverbs and the Pipe (%>%) to turn messy spreadsheets into clean tables without creating intermediate “clutter.”Visualization: Moving beyond basic plots to the layered “Grammar of Graphics” in
ggplot2.Reproducibility: Learning how to write robust functions and manage project scripts.
2. What’s Next?
Today was about the mechanics of R. In the next stage, we move toward interpretation.
Statistics in R Workshop (Part 2)
Data Wrangling: Advanced joining of datasets (merging clinical data with genomic counts).
Statistical Analysis: Performing T-tests, ANOVA, and correlation studies.
Regression: Building linear models to understand the relationship between variables.
Reporting: Using Quarto to create automated PDF or HTML reports for your PI.
3. Resources for Continued R Learning
As you continue your R study, these resources will be your best friends:
Codecademy: Learn R Free Course: Good for those who prefer an interactive, “in-browser” coding experience to practice syntax.
McGill CDSI Workshops: Our colleagues at the Computational and Data Systems Institute offer excellent deep-dives into specific data topics.
McGill QLS-MiCM Workshops: Keep an eye on future MiCM workshops for specialized bioinformatics and high-performance computing training.
4. More workshops
https://www.mcgill.ca/micm/training/workshops-series
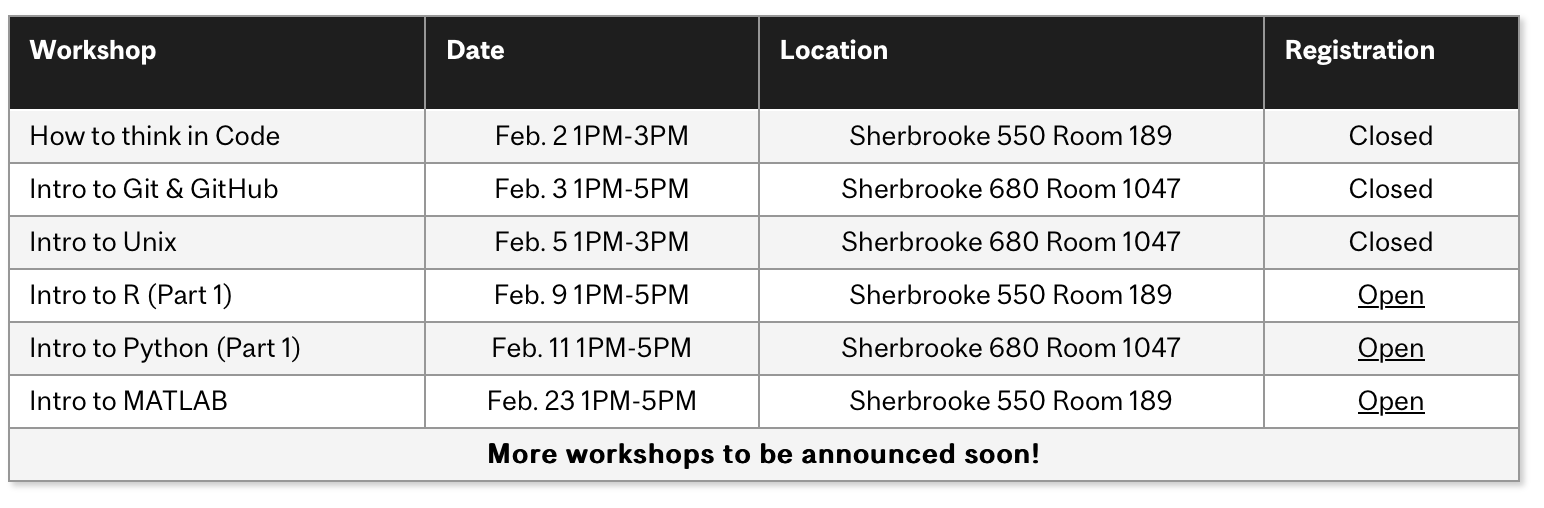
The following is a list of QLS-MiCM workshops offered in the previous year:
How to think in Code
Intro to Git & GitHub
Intro to Unix
Intro to R (Part 1)
Statistics in R (Part 2)
Intro to Python (Part 1)
Intermediate Python (Part 2)
Intro to MATLAB
RNA-seq Bootcamp (DNA2RNA || CDSI)
Data Processing in Python
Intro to Machine Learning
Intro to Molecular Simulations
Proteogenomics
Data Processing for Genetics
Polygenic Risk Scores
5. Thank you for attending! & Feedback Form
You are welcome to scan the QR code below or just click here to share your feedback on today’s workshop. Any suggestions for improvement are warmly welcome!

