# prepare demo dataset
set.seed(228)
demo_data <- data.frame(
"Treatment" = c(rep("ctrl", 6), rep("experiment", 6)),
"Sex" = c(rep("Male", 3), rep("Female", 3), rep("Male", 3), rep("Female", 3)),
"Measurement" = c(sample(100:300, 12))
)
# Calculating means manually
Ctrl_M <- demo_data[which(demo_data$Treatment == "ctrl" & demo_data$Sex == "Male"), "Measurement"]
mean(Ctrl_M)
## [1] 213.3333
Ctrl_F <- demo_data[which(demo_data$Treatment == "ctrl" & demo_data$Sex == "Female"), "Measurement"]
mean(Ctrl_F)
## [1] 149.3333Advanced Data Manipulation
0. The “Base R” Struggle (A Scenario)
Imagine you have data from an experiment with a Control group and an Experimental group. Each group has 3 males and 3 females. You need to calculate the mean for each unique combination (Sex + Treatment).
The “Old” Way (Base R): You have to manually subset the data for every single combination, creating new variables each time.
Now imagine doing this for 10 treatments and 3 timepoints… you would have 60 variables!
There are packages that provide functions to streamline common operations on tabular data and make the code look nicer and cleaner. These packages are part of a broader family called tidyverse, for more information you can visit https://www.tidyverse.org/.
We will cover 5 of the most commonly used functions and combine them using pipes (%>%):
1. select() - used to extract data
2. filter() - to filter entries using logical vectors
3. group_by() - to solve the split-apply-combine problem
4. summarize() - to obtain summary statistics
5. mutate() - to create new columns
1. The tidyverse Philosophy
To solve this “variable explosion,” we use dplyr. It is part of the tidyverse, a collection of R packages that are designed for data science and share a common grammar. The goal is to provide a consistent way to handle data frames. Instead of creating 60 intermediate objects, we create one Pipeline.
if (!require("gapminder", quietly = TRUE))
install.packages("gapminder")
library(gapminder)
if (!require("dplyr", quietly = TRUE))
install.packages("dplyr")
library(dplyr)The Power of the Pipe (%>%)
The pipe operator is the secret sauce of dplyr. It takes the output of one function and “pipes” it directly into the next.
- Think of it as the word “THEN”.
Why use the pipe?
Readability: Standard R code is nested like an onion:
f(g(h(x))). You have to read from the inside out. With the pipe, you read from left to right (or top to bottom).No “Intermediate Clutter”: You don’t have to save
temp_data_1,temp_data_2, etc.Easy Debugging: You can comment out one line of the pipe to see where the data “breaks.”
Comparisons: The Pipe vs. The Nest
The Nest (Hard to read):
# Hard to tell which argument belongs to which function
set.seed(228)
demo_data <- data.frame(
"Treatment" = c(rep("ctrl", 6), rep("experiment", 6)),
"Sex" = c(rep("Male", 3), rep("Female", 3), rep("Male", 3), rep("Female", 3)),
"Measurement" = c(sample(100:300, 12))
)
as.data.frame(summarise(dplyr::group_by(demo_data, Treatment, Sex), Mean = mean(Measurement))) Treatment Sex Mean
1 ctrl Female 149.3333
2 ctrl Male 213.3333
3 experiment Female 167.0000
4 experiment Male 136.0000The Pipe (Easy to read):
# Step-by-step logic
demo_data %>%
dplyr::group_by(Treatment, Sex) %>%
summarise(
Mean = mean(Measurement),
SD = sd(Measurement),
.groups = "drop"
) %>%
as.data.frame() Treatment Sex Mean SD
1 ctrl Female 149.3333 11.06044
2 ctrl Male 213.3333 60.45108
3 experiment Female 167.0000 19.46792
4 experiment Male 136.0000 24.75884Keyboard Shortcut:
Windows/Linux:
Ctrl + Shift + MMac:
Cmd + Shift + M
Fact: The pipe introduced in the
magrittrpackage (2014) and popularized bydplyr(magrittrpackage was named after the artist René Magritte, famous for the “This is not a pipe” painting).Modern R: Since R 4.1.0, there is a built-in pipe
|>. It works similarly but has a few more restrictions (like requiring parentheses after every function).
2. The Core dplyr Verbs
2.1 select(): Picking Columns
Use select() when you only want specific variables (columns) from your dataset. Think of it as a logical sieve: you define the criteria, and only the rows that meet those conditions pass through.
# Select specific columns
gapminder %>%
dplyr::select(year, country, gdpPercap) %>%
head()
## # A tibble: 6 × 3
## year country gdpPercap
## <int> <fct> <dbl>
## 1 1952 Afghanistan 779.
## 2 1957 Afghanistan 821.
## 3 1962 Afghanistan 853.
## 4 1967 Afghanistan 836.
## 5 1972 Afghanistan 740.
## 6 1977 Afghanistan 786.
# Remove a column using the minus sign
gapminder %>%
dplyr::select(-continent) %>%
head()
## # A tibble: 6 × 5
## country year lifeExp pop gdpPercap
## <fct> <int> <dbl> <int> <dbl>
## 1 Afghanistan 1952 28.8 8425333 779.
## 2 Afghanistan 1957 30.3 9240934 821.
## 3 Afghanistan 1962 32.0 10267083 853.
## 4 Afghanistan 1967 34.0 11537966 836.
## 5 Afghanistan 1972 36.1 13079460 740.
## 6 Afghanistan 1977 38.4 14880372 786.Another way of coding:
dplyr::select(.data = gapminder,
year, country, gdpPercap) %>%
head()# A tibble: 6 × 3
year country gdpPercap
<int> <fct> <dbl>
1 1952 Afghanistan 779.
2 1957 Afghanistan 821.
3 1962 Afghanistan 853.
4 1967 Afghanistan 836.
5 1972 Afghanistan 740.
6 1977 Afghanistan 786.dplyr::select(.data = gapminder,
-continent) %>%
head()# A tibble: 6 × 5
country year lifeExp pop gdpPercap
<fct> <int> <dbl> <int> <dbl>
1 Afghanistan 1952 28.8 8425333 779.
2 Afghanistan 1957 30.3 9240934 821.
3 Afghanistan 1962 32.0 10267083 853.
4 Afghanistan 1967 34.0 11537966 836.
5 Afghanistan 1972 36.1 13079460 740.
6 Afghanistan 1977 38.4 14880372 786.2.2 filter(): Picking Rows
Use filter() to find observations (rows) that meet a certain condition.
# Include only European countries in the year 2007
gapminder %>%
dplyr::filter(continent == "Europe", year == 2007) %>%
dplyr::select(country, lifeExp)
## # A tibble: 30 × 2
## country lifeExp
## <fct> <dbl>
## 1 Albania 76.4
## 2 Austria 79.8
## 3 Belgium 79.4
## 4 Bosnia and Herzegovina 74.9
## 5 Bulgaria 73.0
## 6 Croatia 75.7
## 7 Czech Republic 76.5
## 8 Denmark 78.3
## 9 Finland 79.3
## 10 France 80.7
## # ℹ 20 more rowsThe Base R Way
The logic is “nested.” You have to read from the inside out to understand what is happening, and you must repeat the name of the data frame (gapminder$) multiple times.
gapminder[which(gapminder$continent == "Europe" & gapminder$year == 2007),
c("country", "lifeExp")]
## # A tibble: 30 × 2
## country lifeExp
## <fct> <dbl>
## 1 Albania 76.4
## 2 Austria 79.8
## 3 Belgium 79.4
## 4 Bosnia and Herzegovina 74.9
## 5 Bulgaria 73.0
## 6 Croatia 75.7
## 7 Czech Republic 76.5
## 8 Denmark 78.3
## 9 Finland 79.3
## 10 France 80.7
## # ℹ 20 more rows3. The Power Duo: group_by() and summarize()
This is the most common workflow in bioinformatics. You split the data into groups, apply a calculation, and combine the results into a summary table.
3.1 group_by()
This doesn’t change the data visually; it creates “hidden” groups that R remembers for the next step.
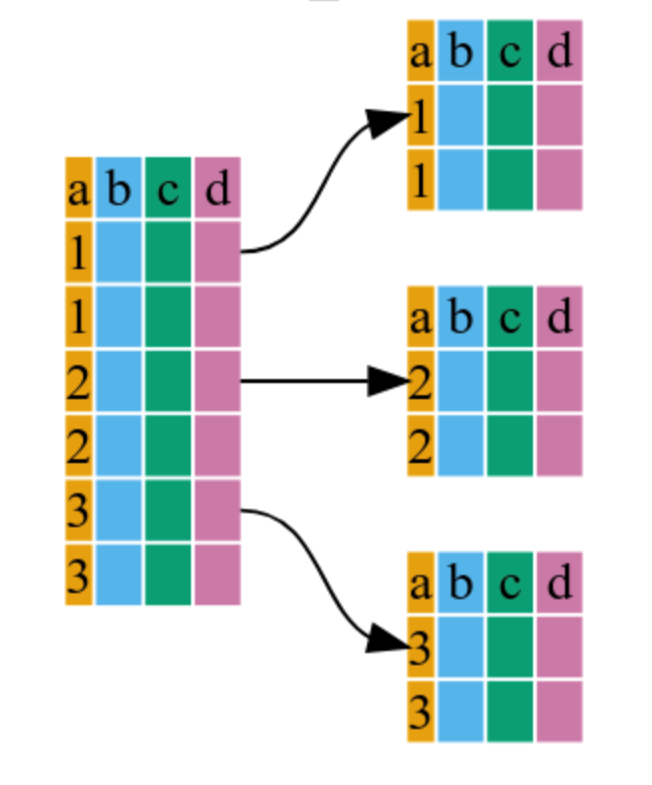
gapminder %>%
dplyr::group_by(continent)
## # A tibble: 1,704 × 6
## # Groups: continent [5]
## country continent year lifeExp pop gdpPercap
## <fct> <fct> <int> <dbl> <int> <dbl>
## 1 Afghanistan Asia 1952 28.8 8425333 779.
## 2 Afghanistan Asia 1957 30.3 9240934 821.
## 3 Afghanistan Asia 1962 32.0 10267083 853.
## 4 Afghanistan Asia 1967 34.0 11537966 836.
## 5 Afghanistan Asia 1972 36.1 13079460 740.
## 6 Afghanistan Asia 1977 38.4 14880372 786.
## 7 Afghanistan Asia 1982 39.9 12881816 978.
## 8 Afghanistan Asia 1987 40.8 13867957 852.
## 9 Afghanistan Asia 1992 41.7 16317921 649.
## 10 Afghanistan Asia 1997 41.8 22227415 635.
## # ℹ 1,694 more rows3.2 summarize()
This “collapses” each group into a single row of statistics.
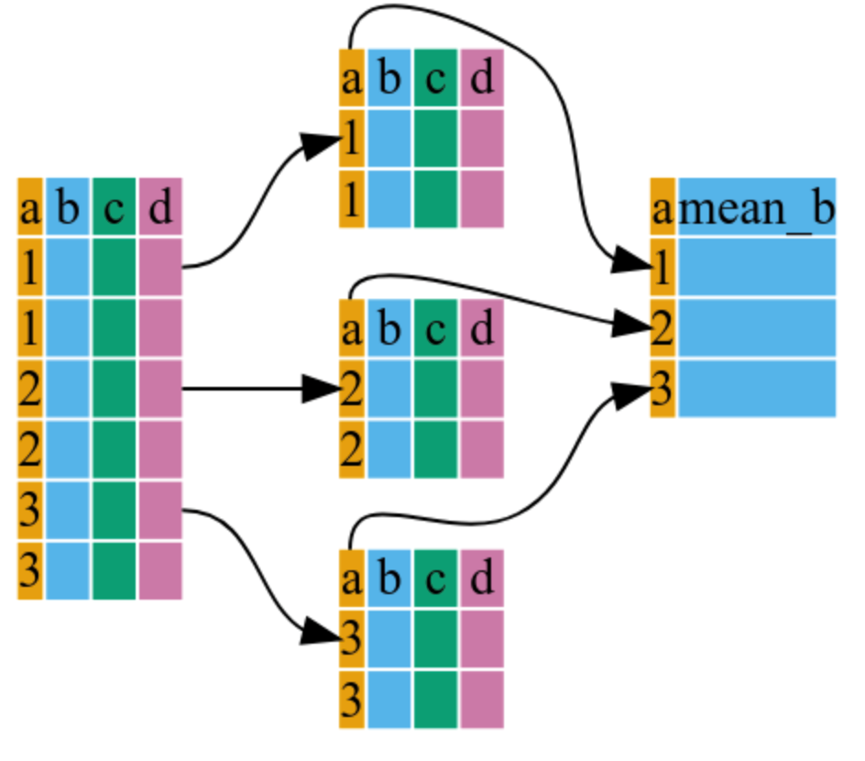
# Calculate mean GDP and Standard Error for each continent
gapminder %>%
dplyr::group_by(continent) %>%
dplyr::summarize(
mean_le = mean(lifeExp),
min_le = min(lifeExp),
max_le = max(lifeExp),
se_le = sd(lifeExp) / sqrt(dplyr::n()) # n() counts the number of observations
)
## # A tibble: 5 × 5
## continent mean_le min_le max_le se_le
## <fct> <dbl> <dbl> <dbl> <dbl>
## 1 Africa 48.9 23.6 76.4 0.366
## 2 Americas 64.7 37.6 80.7 0.540
## 3 Asia 60.1 28.8 82.6 0.596
## 4 Europe 71.9 43.6 81.8 0.286
## 5 Oceania 74.3 69.1 81.2 0.7754. mutate(): Creating New Variables
mutate() allows you to add new columns while keeping the old ones. It is perfect for calculations like unit conversions or normalization.
# Create a new column for GDP in billions
gapminder %>%
dplyr::mutate(gdp_billion = gdpPercap * pop / 10^9) %>%
head()
## # A tibble: 6 × 7
## country continent year lifeExp pop gdpPercap gdp_billion
## <fct> <fct> <int> <dbl> <int> <dbl> <dbl>
## 1 Afghanistan Asia 1952 28.8 8425333 779. 6.57
## 2 Afghanistan Asia 1957 30.3 9240934 821. 7.59
## 3 Afghanistan Asia 1962 32.0 10267083 853. 8.76
## 4 Afghanistan Asia 1967 34.0 11537966 836. 9.65
## 5 Afghanistan Asia 1972 36.1 13079460 740. 9.68
## 6 Afghanistan Asia 1977 38.4 14880372 786. 11.75. Putting It All Together
The true beauty of dplyr is chaining everything into one clean pipeline.
# A complete pipeline: Create a variable, group it, and summarize it
summary_table <- gapminder %>%
dplyr::mutate(gdp_billion = gdpPercap * pop / 10^9) %>%
dplyr::group_by(continent, year) %>%
dplyr::summarize(
mean_gdpPercap = mean(gdpPercap),
sd_gdpPercap = sd(gdpPercap),
mean_pop = mean(pop),
sd_pop = sd(pop),
mean_gdp_billion = mean(gdp_billion),
sd_gdp_billion = sd(gdp_billion)
)
head(summary_table)
## # A tibble: 6 × 8
## # Groups: continent [1]
## continent year mean_gdpPercap sd_gdpPercap mean_pop sd_pop mean_gdp_billion
## <fct> <int> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 Africa 1952 1253. 983. 4570010. 6.32e6 5.99
## 2 Africa 1957 1385. 1135. 5093033. 7.08e6 7.36
## 3 Africa 1962 1598. 1462. 5702247. 7.96e6 8.78
## 4 Africa 1967 2050. 2848. 6447875. 8.99e6 11.4
## 5 Africa 1972 2340. 3287. 7305376. 1.01e7 15.1
## 6 Africa 1977 2586. 4142. 8328097. 1.16e7 18.7
## # ℹ 1 more variable: sd_gdp_billion <dbl>dplyr::?
You’ll notice I used dplyr::select() instead of just select(). As we discussed in the “Functions” module, many packages have a filter or select function. Being explicit ensures that your code never breaks, even if you load other libraries later.
